拳皇十周年纪念版无限气下载
alicucu 2026-01-19 10:32 2 浏览
下载一个啪啪模拟器在里面找有很多拳皇包括十周年而且里面有干枯大地七枷社
浏览器搜索游戏名、打开相关网站的网页、可以下载游戏安装文件!!
1.
以win10,谷歌浏览器为例。
打开谷歌浏览器

2.
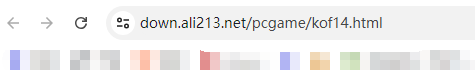
浏览器地址栏输入网址https://down.ali213.net/pcgame/kof14.html
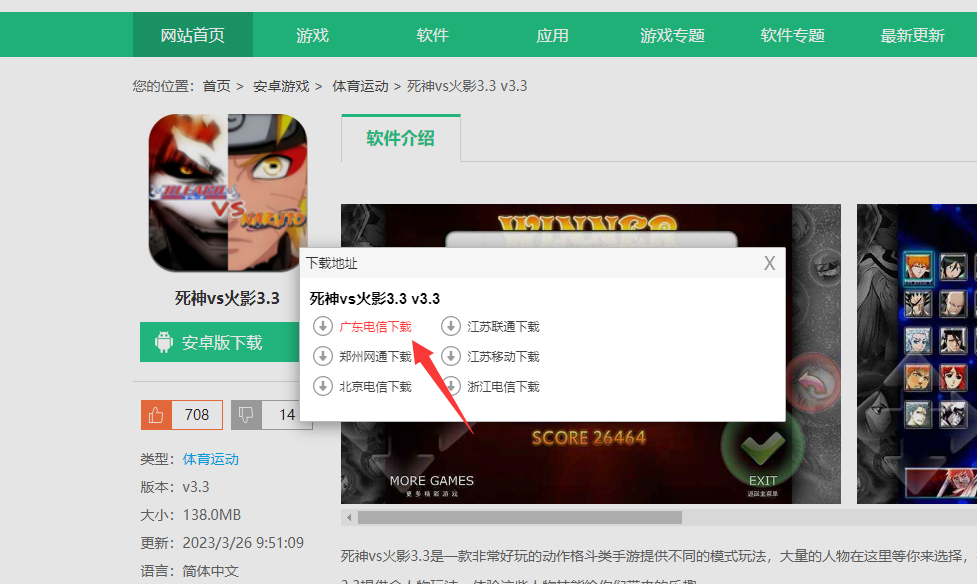
3.
点击资源检索

4.
点击电信高速下载
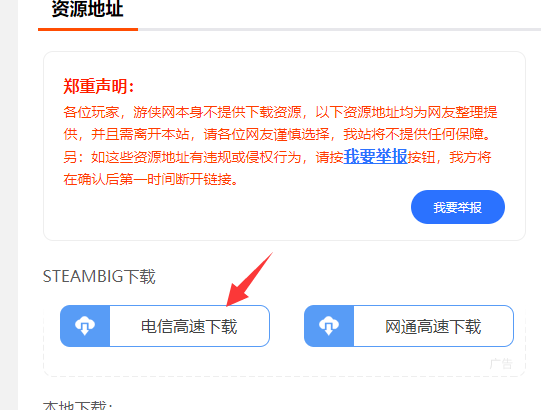
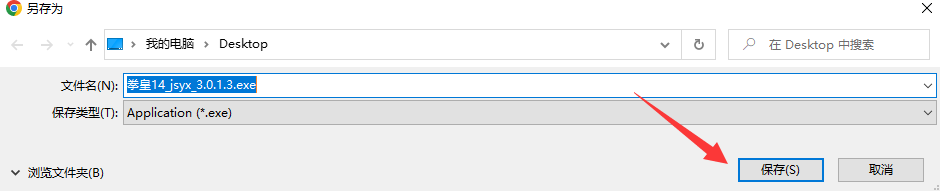
5.
点击保存即可
- 上一篇:魔兽争霸3下载官方(魔兽争霸3下载官方网站)
- 下一篇:使命召唤16单机版中文破解版
相关推荐
- 怪物猎人崛起官网(怪物猎人崛起官网下载)
-
《怪物猎人:崛起》体验版现已在NintendoeShop上再次发布除了上次的体验版中包含的全部14种武器之基本操作和实战任务之外,此次的体验版还新增主题魔物“怨虎龙”的讨伐任务。追加怨虎龙讨伐任务!...
- 暗黑2十大变态技能(暗黑2十大变态技能是什么)
-
近战型PVC武学龙系刺客: 特点:攻防一体。近身格斗型,不怕物免,不怕铁娘子,操作简单安全。由于使用的攻击技能都不需要很高IAS的支持,所以双手选择的余地很多。是一个不错的武学系ASN。 技能...
- 仙剑奇侠手游(仙剑奇侠手游中剁椒鱼头配料)
-
1攻击对象选择:根据阵型的站位,分为3行5列。普通攻击和无特殊指向的技能会优先攻击第1行和自己位于同一列的敌人,如果同列第1行没有敌人,则对象改为离自己最近列前排的目标。2逐次将前排的敌人全部清除,才...
- 使命召唤16单机版中文破解版
-
要下载CallofDuty16单机版,您可以按照以下步骤进行操作:1.打开您的网络浏览器,访问CallofDuty官方网站(https://www.callofduty.com/)...
- 魔兽争霸3下载官方(魔兽争霸3下载官方网站)
-
1、首先下载手游模拟器。2、下载冰封王座电脑版的游戏包。3、游戏包下载完成后点击模拟器右上角的本地安装,添加魔兽世界游戏包即可开始安装游戏。3、选择默认的引擎,点击“确定安装”即可。4、完成安装,玩家...
- 阿卡丽的神秘商店入口网址(阿卡丽的神秘商店入口网址是多少)
-
阿卡丽的神秘商店暂时关闭原因:由于阿卡丽的神秘商店出现BUG,需要暂时关闭。我们的工程师正在紧急修复中。阿卡丽的神秘商店再次开放时间:再次开放时间预计为4月2日1:00。阿卡丽神秘商店是《英雄联盟》游...
- 红色警戒单机版电脑版(红色警戒单机版怎么玩)
-
在红警之家网站上下载一个红警客户端,然后打开客户端安装,按照指示操作,就可以安装完成,并双击桌面上的快捷方式,就可以玩了。1.打开电脑上的任意一款网络浏览器。2.在地址栏输入红警之家官网地址。3.点击...
-

- 手游排行榜前十名单机游戏(手游排行榜前十名单机游戏推荐)
-
我觉得比较好玩的十个手机单机游戏有:《地狱边境》、《刺客信条》、《狂爆之翼》、《阿尔托的冒险》、《滑雪大冒险》、《方舟》、《使命召唤(手游版)》、《我的世界》、《这是我的战争》、《地球末日:生存》等。1、《狂爆之翼》是一款以激爽战斗为核心的...
-
2026-01-19 09:22 alicucu
- 免费体验个10小时的云游戏(云游戏免费秒玩入口)
-
每天能提体现12个钟很好玩的一些很游戏1云游戏体验时间到了后需要重新购买或续费才能继续使用2这是由于云游戏需要连接远程服务器来进行游戏,而服务器使用的是租赁制,一般都会按照时间计费,所以在使用...
- 三国杀十周年(三国杀十周年现在叫什么)
-
氪金消费,手杀和ol最烧钱,经典6k拉满大败而归。神郭嘉时代的6w5(就是6w5抽盒子没出货权1)。ol和手杀想要阴雷这些也要几百。但是十周年像许攸郝昭这些都是白嫖。从价格来说十周年最便宜ui看个人喜...
- lol官网领取中心(lol官方领取中心)
-
英雄联盟官网右下角有一个领取中心。在游戏的领取中心界面往下滑,可以看到一个【可领取的道具】,在这个界面还可以继续往下滑,之后就可以看到一个【永久道具宝箱领取】了。该宝箱可以获得的皮肤有很多,KDA阿狸...
- 地下城与勇士手游版下载安装
-
回答如下:地下城与勇士是一款网络游戏,需要在官方网站或游戏平台下载安装。以下是在官方网站下载安装的步骤:1.打开官方网站:https://dnf.qq.com/,点击“下载游戏”按钮。2.选择“P...
- 一周热门
- 最近发表
- 标签列表
-